Monitor and Check Experiments#
How to check the experiment configuration#
To check the configuration of the experiment, use the command:
autosubmit check EXPID
EXPID is the experiment identifier.
It checks experiment configuration and warns about any detected error or inconsistency. It is used to check if the script is well-formed. If any template has an inconsistency it will replace them for an empty value on the cmd generated. Options:
usage: autosubmit check [-h -nt] expid
expid experiment identifier
-nt --notransitive
prevents doing the transitive reduction when plotting the workflow
-h, --help show this help message and exit
Example:
autosubmit check cxxx
How to use check in running time:#
In jobs_cxxx.yml , you can set check(default true) to check the scripts during autosubmit run cxx.
There are two parameters related to check:
CHECK: Controls the mechanism that allows replacing an unused variable with an empty string ( %_% substitution). It is TRUE by default.
SHOW_CHECK_WARNINGS: For debugging purposes. It will print a lot of information regarding variables and substitution if it is set to TRUE.
CHECK: TRUE or FALSE or ON_SUBMISSION # Default is TRUE
SHOW_CHECK_WARNINGS: TRUE or FALSE # Default is FALSE
CHECK: TRUE # Static templates (existing on `autosubmit create`). Used to substitute empty variables
CHECK: ON_SUBMISSION # Dynamic templates (generated on running time). Used to substitute empty variables.
CHECK: FALSE # Used to disable this substitution.
SHOW_CHECK_WARNINGS: TRUE # Shows a LOT of information. Disabled by default.
For example:
LOCAL_SETUP:
FILE: filepath_that_exists
PLATFORM: LOCAL
WALLCLOCK: 05:00
CHECK: TRUE
SHOW_CHECK_WARNINGS: TRUE
...
SIM:
FILE: filepath_that_no_exists_until_setup_is_processed
PLATFORM: bsc_es
DEPENDENCIES: LOCAL_SETUP SIM-1
RUNNING: chunk
WALLCLOCK: 05:00
CHECK: ON_SUBMISSION
SHOW_CHECK_WARNINGS: FALSE
...
How to generate cmd files#
To generate the cmd files of the current non-active jobs experiment, it is possible to use the command:
autosubmit inspect EXPID
EXPID is the experiment identifier.
Usage#
Options:
usage: autosubmit inspect [-h] [-fl] [-fc] [-fs] [-ft] [-cw] expid
expid experiment identifier
-h, --help show this help message and exit
-fl FILTER_LIST, --list
List of job names to be generated
-fc FILTER_CHUNK, --filter_chunk
List of chunks to be generated
-fs FILTER_STATUS, --filter_status
List of status to be generated
-ft FILTER_TYPE, --filter_type
List of types to be generated
-cw --checkwrapper
Generate the wrapper cmd with the current filtered jobs
-f --force
Generate all cmd files
Example#
with autosubmit.lock present or not:
autosubmit inspect expid
with autosubmit.lock present or not:
autosubmit inspect expid -f
without autosubmit.lock:
autosubmit inspect expid -fl [-fc,-fs or ft]
To generate cmd for wrappers:
autosubmit inspect expid -cw -f
With autosubmit.lock and no (-f) force, it will only generate all files that are not submitted.
Without autosubmit.lock, it will generate all unless filtered by -fl,fc,fs or ft.
To generate cmd only for a single job of the section :
autosubmit inspect expid -q
How to monitor an experiment#
To monitor the status of the experiment, use the command:
autosubmit monitor EXPID
EXPID is the experiment identifier.
Options:
usage: autosubmit monitor [-h] [-o {pdf,png,ps,svg,txt}] [-group_by {date,member,chunk,split} -expand -expand_status] [-fl] [-fc] [-fs] [-ft] [-cw] expid [-txt] [-txtlog]
expid Experiment identifier.
-h, --help Show this help message and exit.
-o {pdf,png,ps,svg}, --output {pdf,png,ps,svg,txt}
Type of output for generated plot (or text file).
-group_by {date,member,chunk,split,automatic}
Criteria to use for grouping jobs.
-expand, List of dates/members/chunks to expand.
-expand_status, Status(es) to expand.
-fl FILTER_LIST, --list
List of job names to be filtered.
-fc FILTER_CHUNK, --filter_chunk
List of chunks to be filtered.
-fs FILTER_STATUS, --filter_status
Status to be filtered.
-ft FILTER_TYPE, --filter_type
Type to be filtered.
--hide, Hide the plot.
-txt, --text
Generates a tree view format that includes job name, children number, and status in a file in the /status/ folder. If possible, shows the results in the terminal.
-txtlog, --txt_logfiles
Generates a list of job names, status, .out path, and .err path as a file in /status/ (AS <3.12 behaviour).
-nt --notransitive
Prevents doing the transitive reduction when plotting the workflow.
-cw --check_wrapper
Generate the wrapper in the current workflow.
Example:
autosubmit monitor cxxx
The location where the user can find the generated plots with date and timestamp can be found below:
<experiments_directory>/cxxx/plot/cxxx_<date>_<time>.pdf
The location where the user can find the txt output containing the status of each job and the path to out and err log files.
<experiments_directory>/cxxx/status/cxxx_<date>_<time>.txt
Hint
Very large plots may be a problem for some pdf and image viewers. If you are having trouble with your usual monitoring tool, try using svg output and opening it with Google Chrome with the SVG Navigator extension installed.
In order to understand more the grouping options, please check Grouping jobs.
Grouping jobs#
Other than the filters, another option for large workflows is to group jobs. This option is available with the group_by keyword, which can receive the values {date,member,chunk,split,automatic}.
For the first 4 options, the grouping criteria is explicitly defined {date,member,chunk,split}.
In addition to that, it is possible to expand some dates/members/chunks that would be grouped either/both by status or/and by specifying the date/member/chunk not to group.
The syntax used in this option is almost the same as for the filters, in the format of [ date1 [ member1 [ chunk1 chunk2 ] member2 [ chunk3 ... ] ... ] date2 [ member3 [ chunk1 ] ] ... ]
Important
The grouping option is also in autosubmit monitor, create, setstatus and recovery
Examples:
Consider the following workflow:
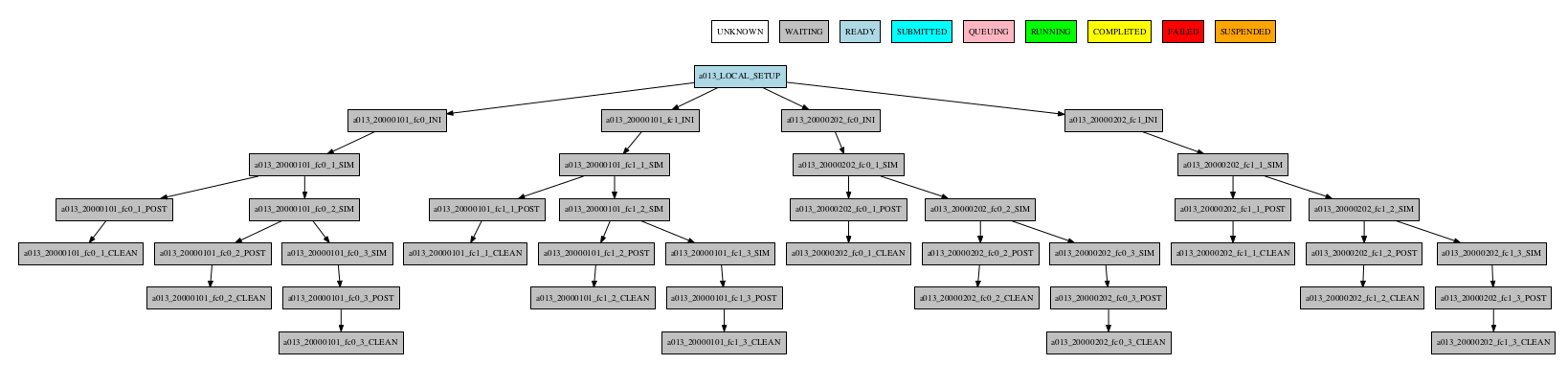
Group by date
-group_by=date
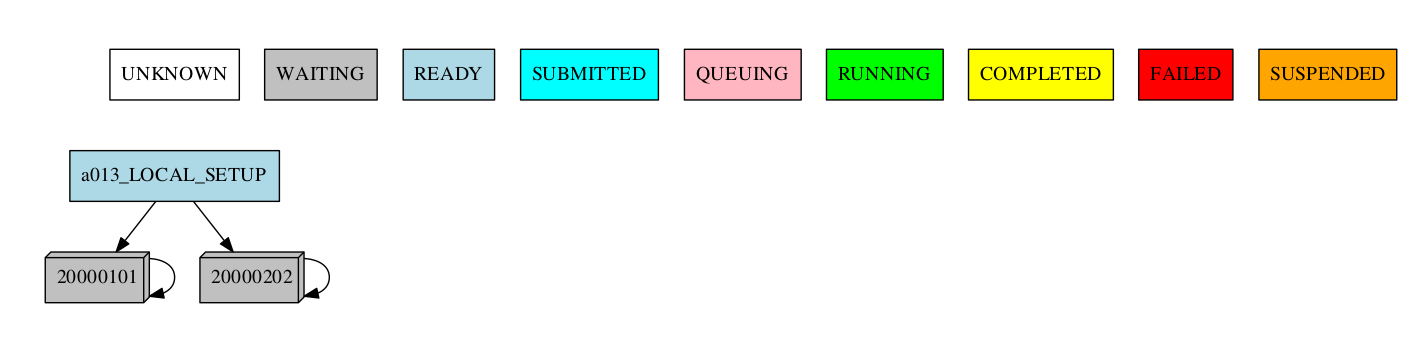
-group_by=date -expand="[ 20000101 ]"
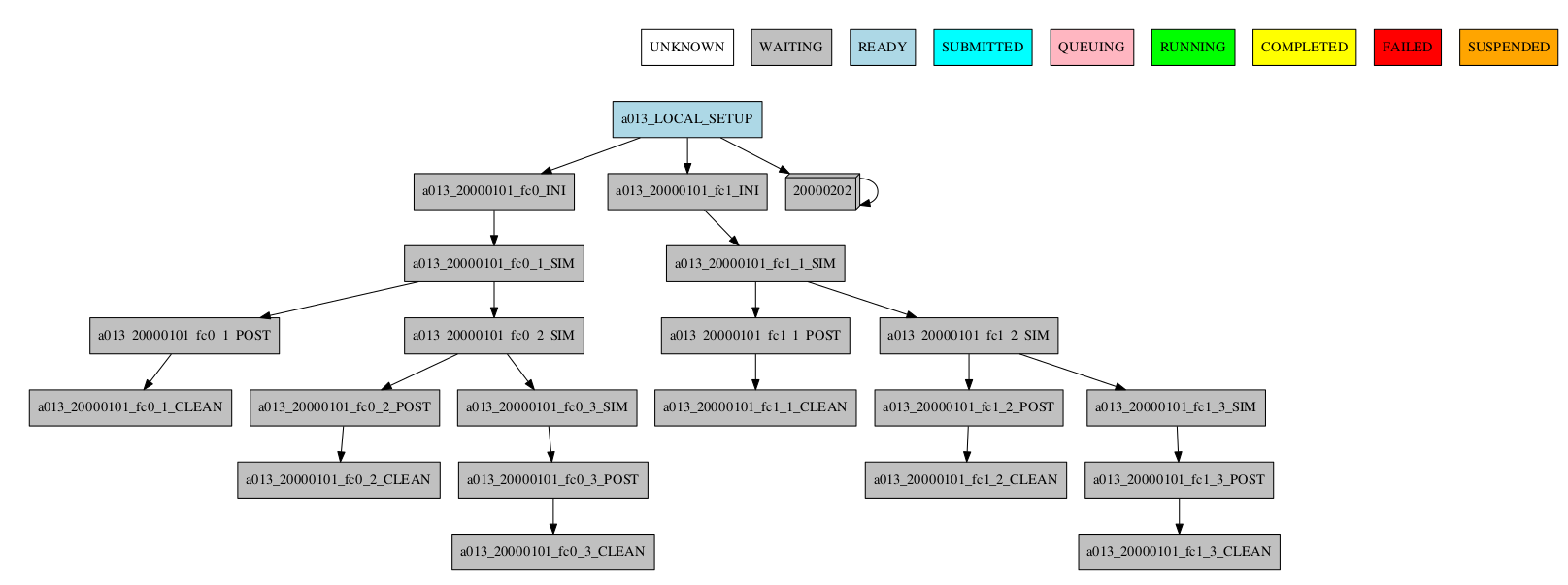
-group_by=date -expand_status="FAILED RUNNING"
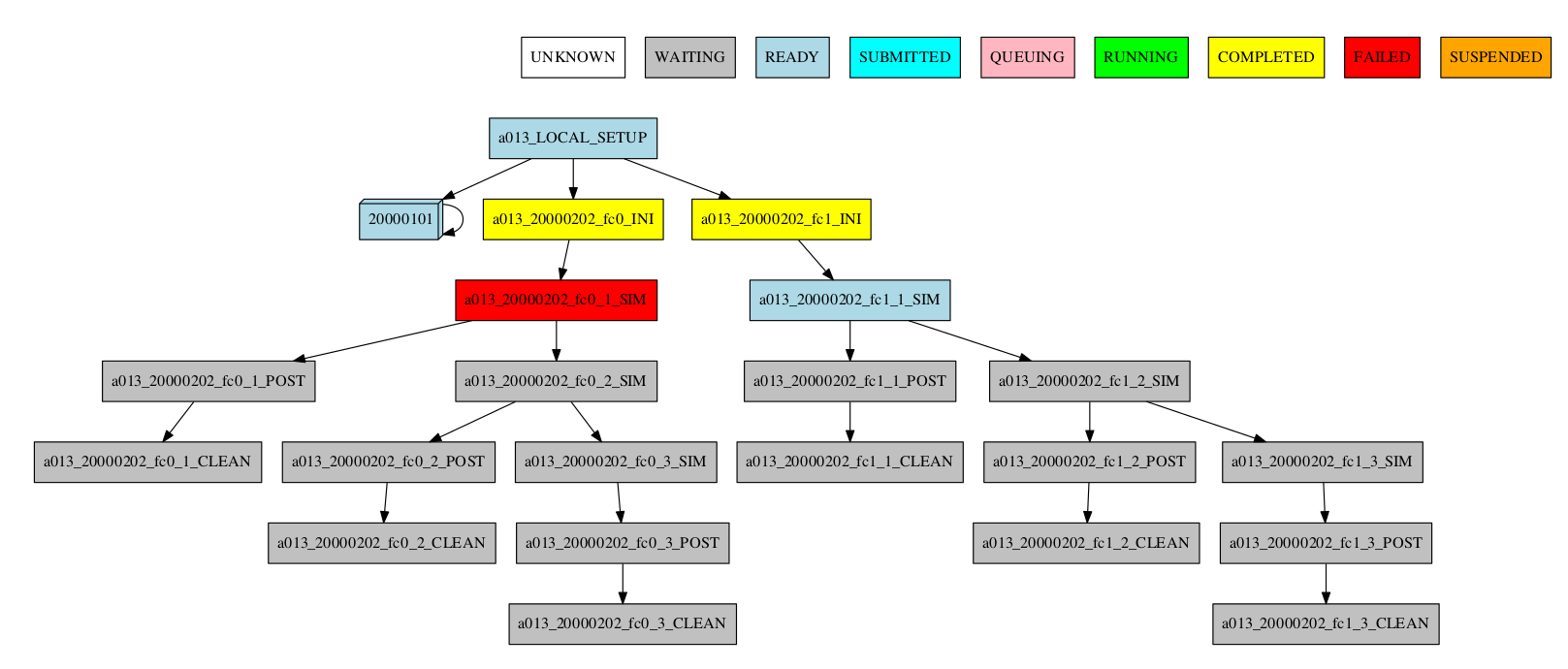
-group_by=date -expand="[ 20000101 ]" -expand_status="FAILED RUNNING"
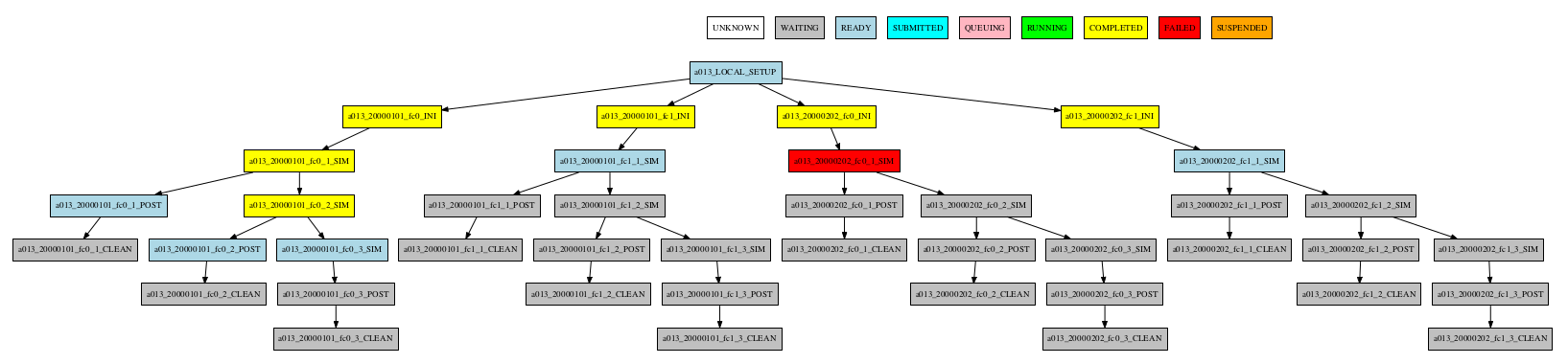
Group by member
-group_by=member
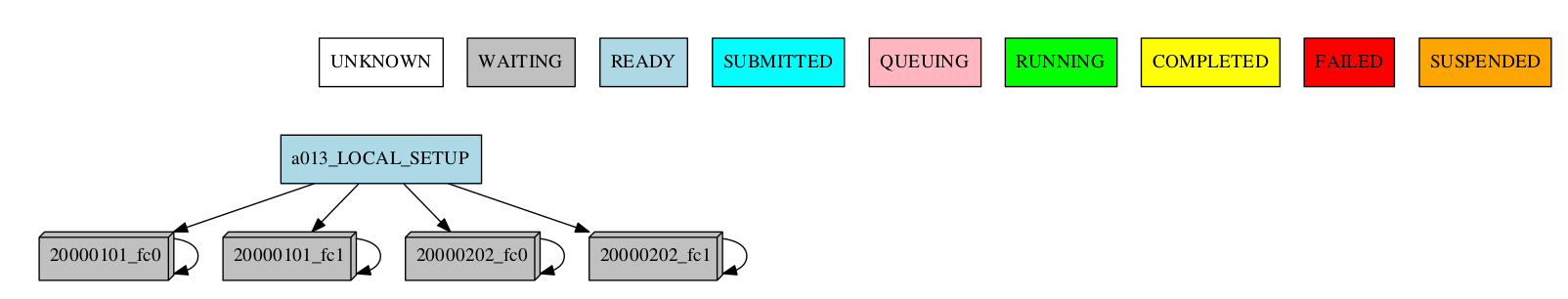
-group_by=member -expand="[ 20000101 [ fc0 fc1 ] 20000202 [ fc0 ] ]"
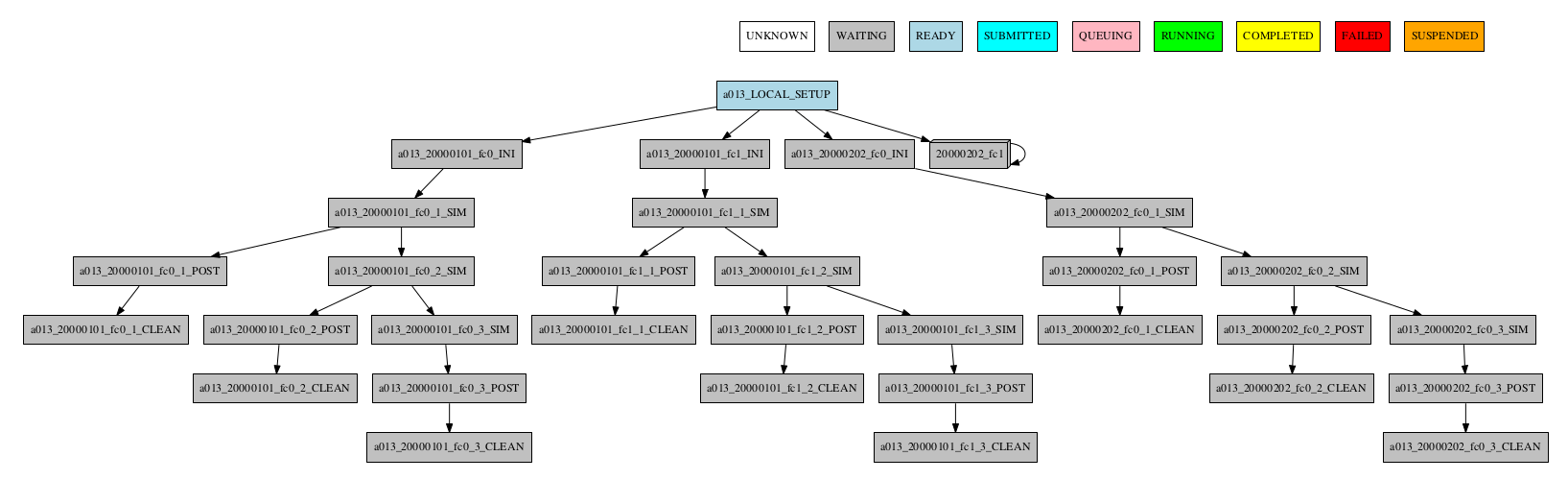
-group_by=member -expand_status="FAILED QUEUING"
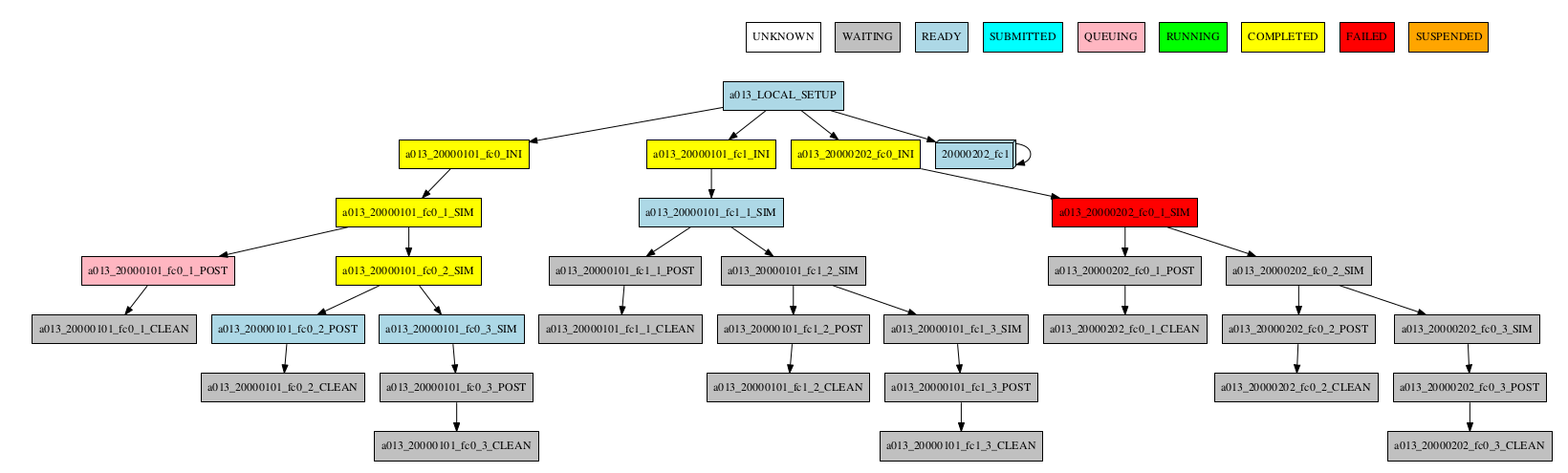
-group_by=member -expand="[ 20000101 [ fc0 fc1 ] 20000202 [ fc0 ] ]" -expand_status="FAILED QUEUING"
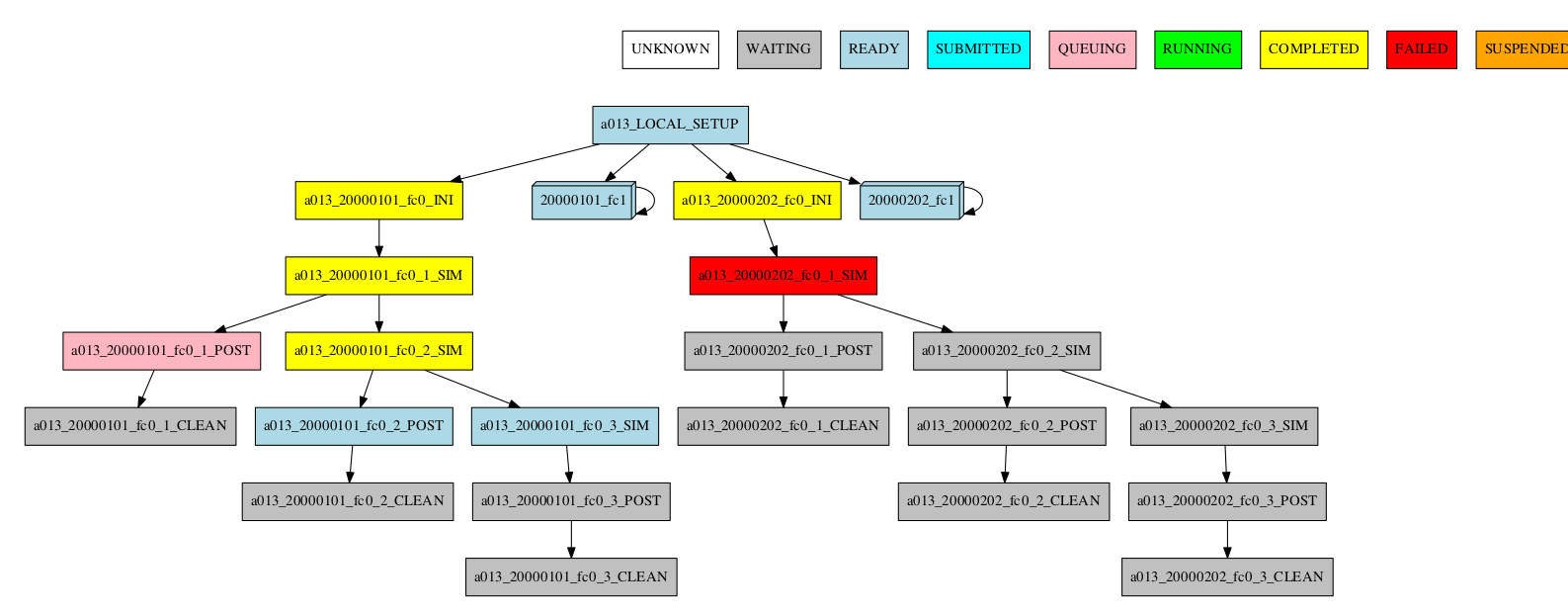
Group by chunk
-group_by=chunk
TODO: Add group_chunk.png figure.
Synchronize jobs
If there are jobs synchronized between members or dates, then a connection between groups is shown:
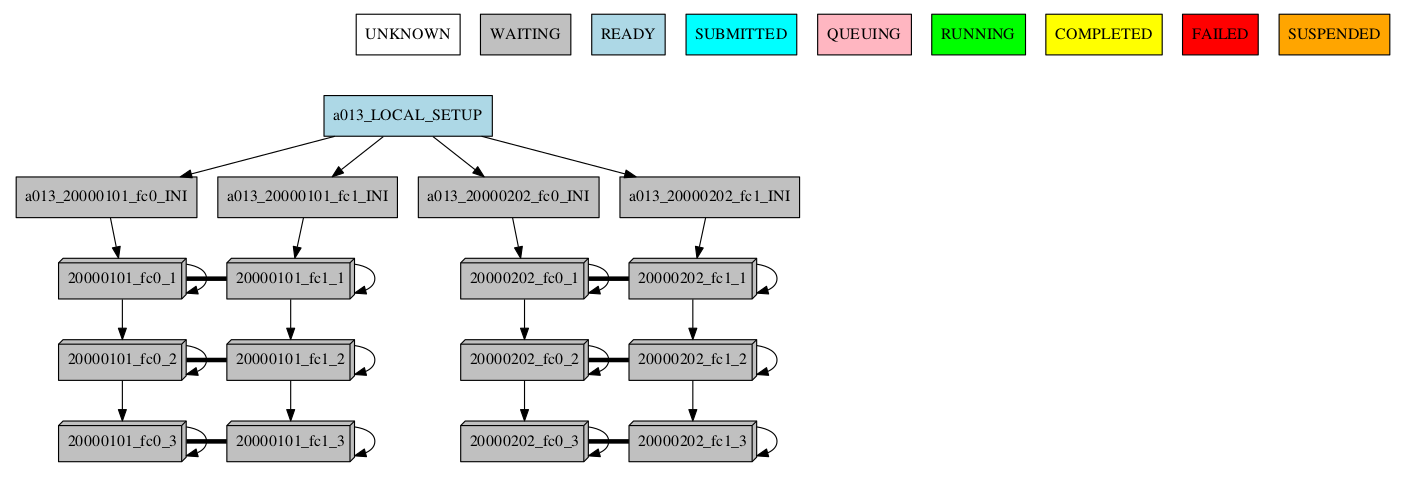
-group_by=chunk -expand="[ 20000101 [ fc0 [1 2] ] 20000202 [ fc1 [2] ] ]"
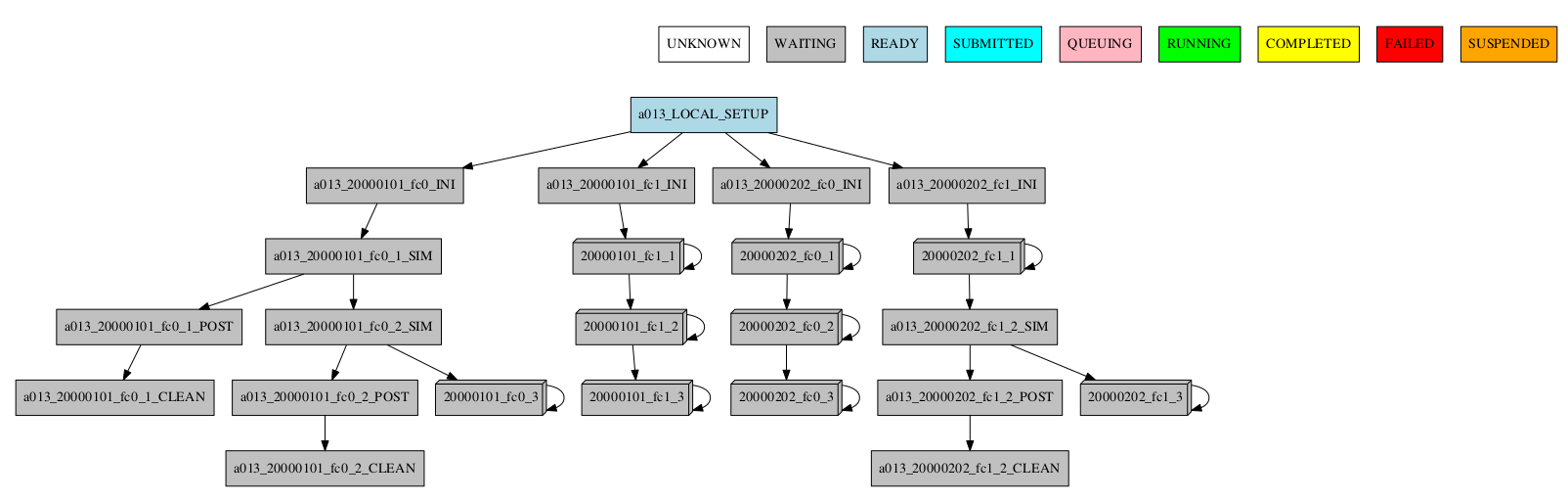
-group_by=chunk -expand_status="FAILED RUNNING"
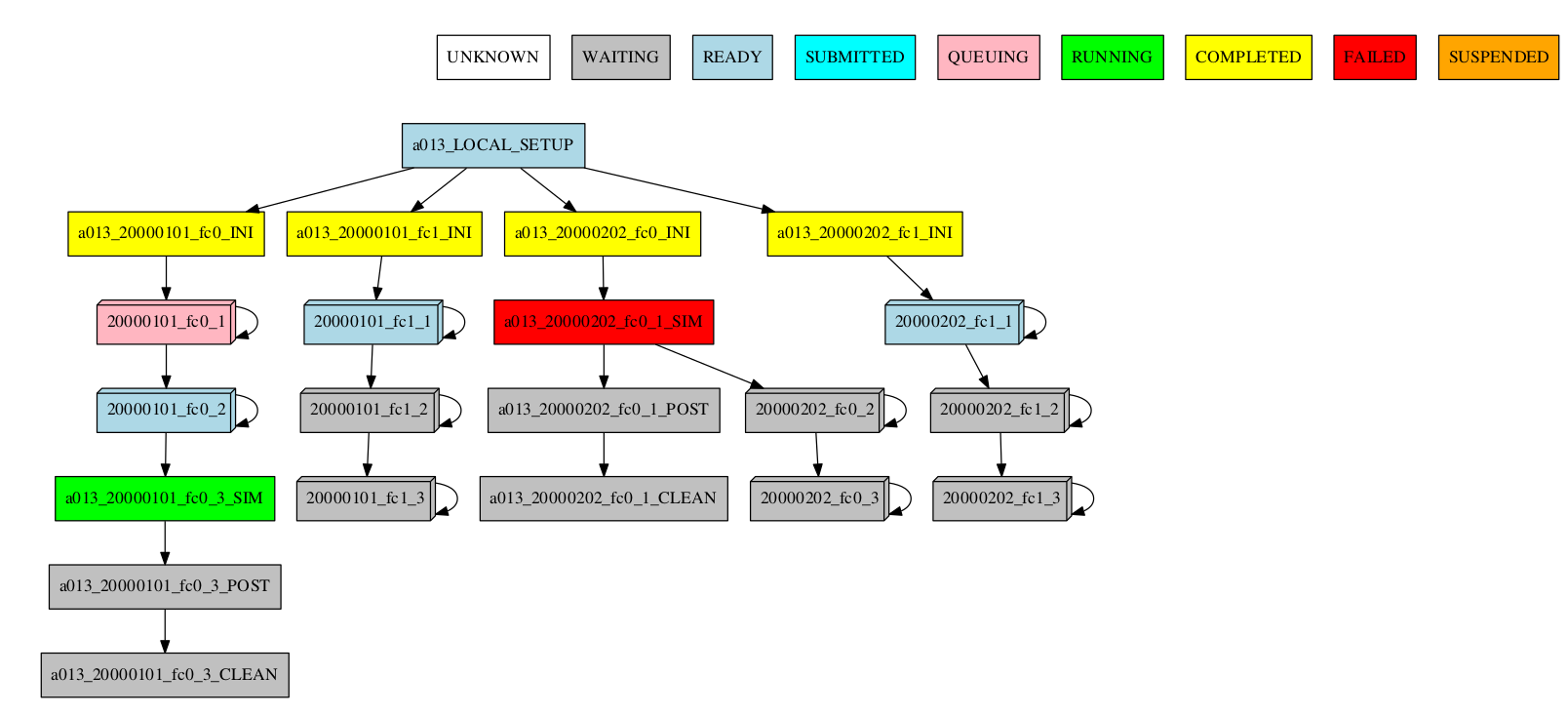
-group_by=chunk -expand="[ 20000101 [ fc0 [1] ] 20000202 [ fc1 [1 2] ] ]" -expand_status="FAILED RUNNING"
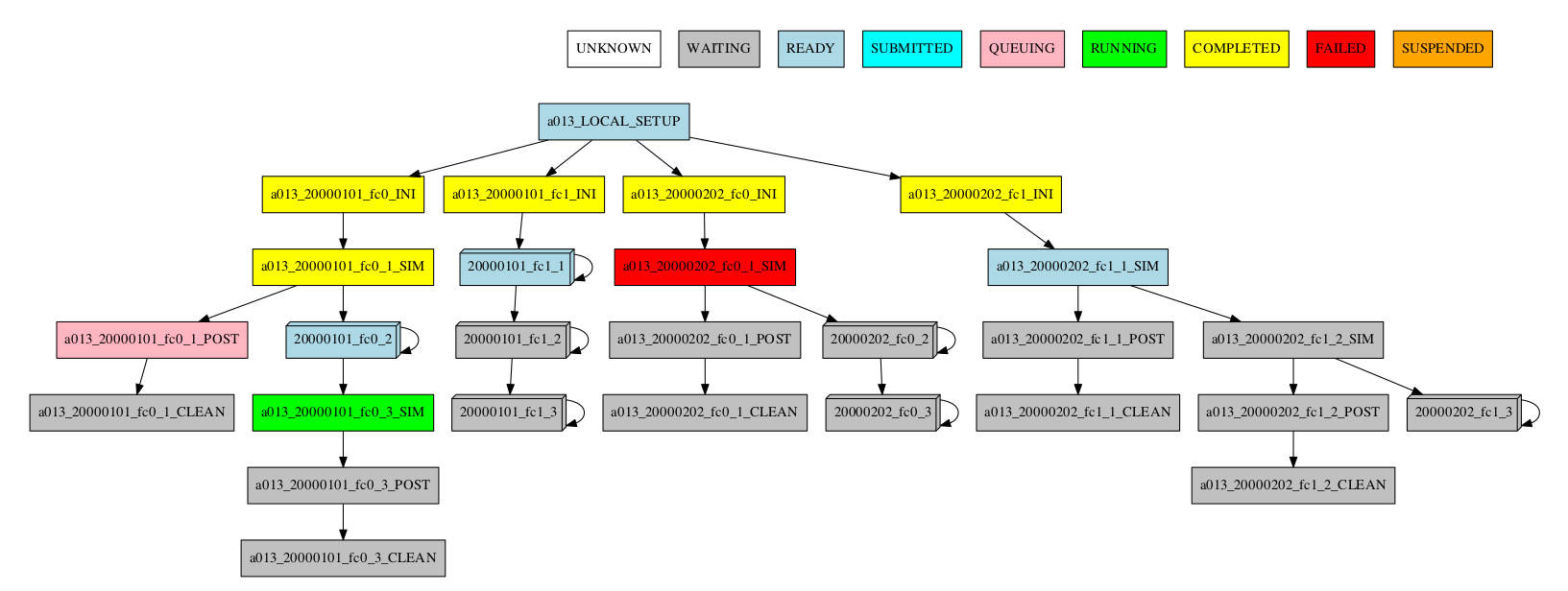
Group by split
If there are chunk jobs that are split, the splits can also be grouped.
-group_by=split

Understanding the group status
If there are jobs with different status grouped together, the status of the group is determined as follows: If there is at least one job that failed, the status of the group will be FAILED. If there are no failures, but if there is at least one job running, the status will be RUNNING. The same idea applies following the hierarchy: SUBMITTED, QUEUING, READY, WAITING, SUSPENDED, UNKNOWN. If the group status is COMPLETED, it means that all jobs in the group were completed.
Automatic grouping
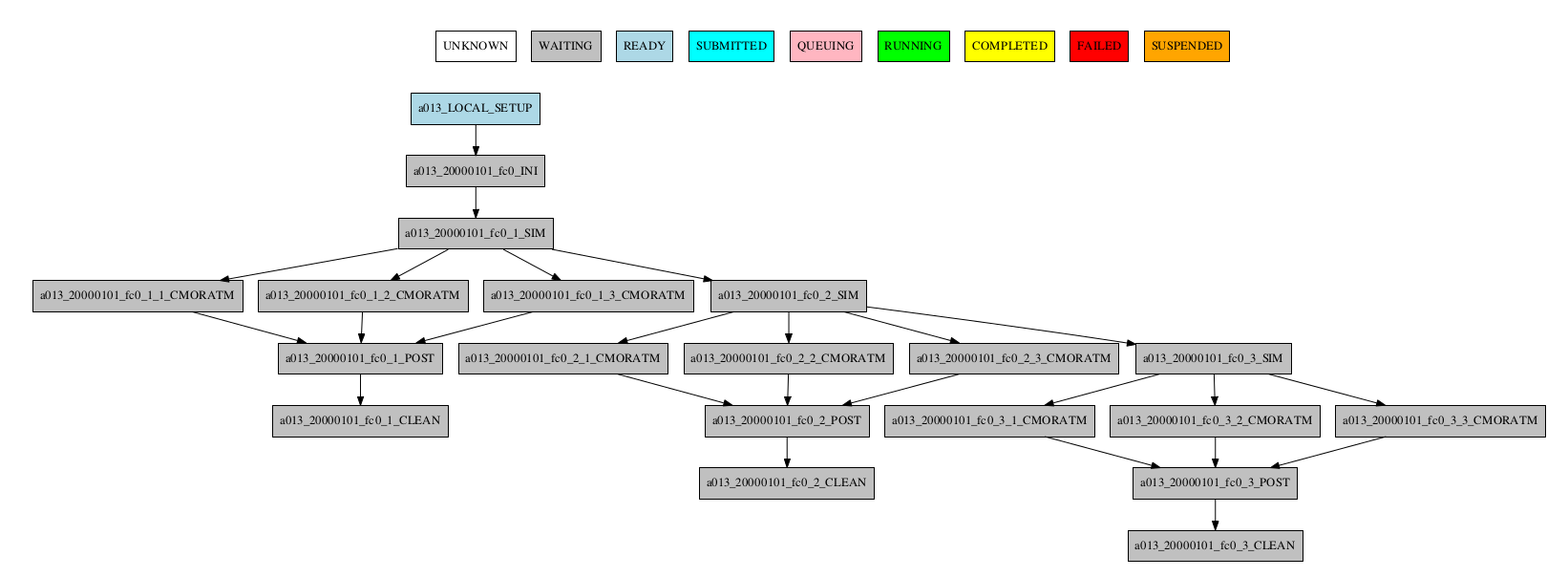
For the automatic grouping, the groups are created by collapsing the split->chunk->member->date that share the same status (following this hierarchy). The following workflow automatic created the groups 20000101_fc0, since all the jobs for this date and member were completed, 20000101_fc1_3, 20000202_fc0_2, 20000202_fc0_3 and 20000202_fc1, as all the jobs up to the respective group granularity share the same - waiting - status.
For example:
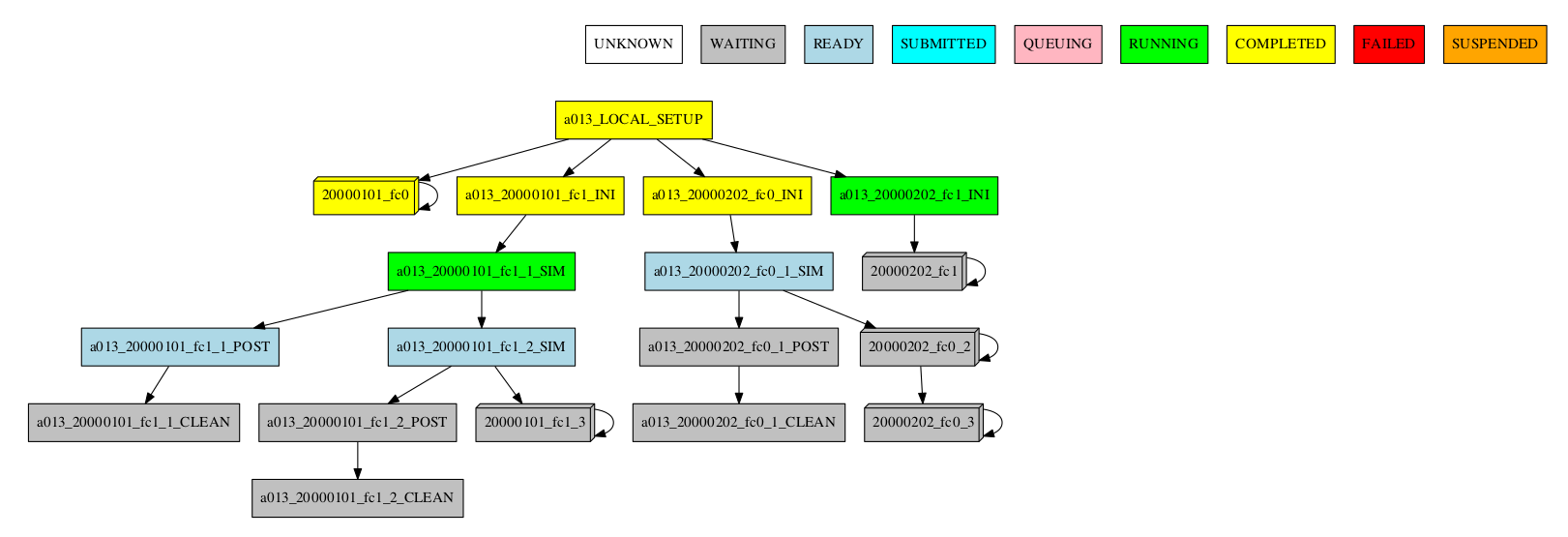
Especially in the case of monitoring an experiment with a very large number of chunks, it might be useful to hide the groups created automatically. This allows to better visualize the chunks in which there are jobs with different status, which can be a good indication that there is something currently happening within such chunks (jobs ready, submitted, running, queueing or failed).
-group_by=automatic --hide_groups
How to profile Autosubmit while monitoring an experiment#
Autosubmit offers the possibility to profile the execution of the monitoring process. To enable the
profiler, just add the --profile (or -p) flag to your autosubmit monitor command, as in
the following example:
autosubmit monitor --profile EXPID
Note
Remember that the purpose of this profiler is to measure the performance of Autosubmit, not the jobs it runs.
This profiler uses Python’s cProfile and psutil modules to generate a report with simple CPU and
memory metrics which will be displayed in your console after the command finishes, as in the example below:
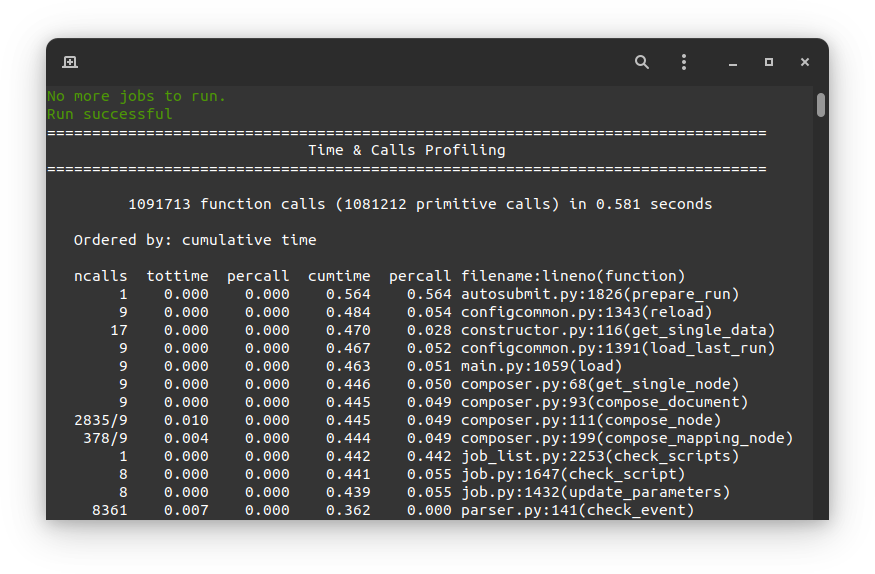
The profiler output is also saved in <EXPID>/tmp/profile. There you will find two files, the
report in plain text format and a .prof binary which contains the CPU metrics. We highly recommend
using SnakeViz to visualize this file, as follows:
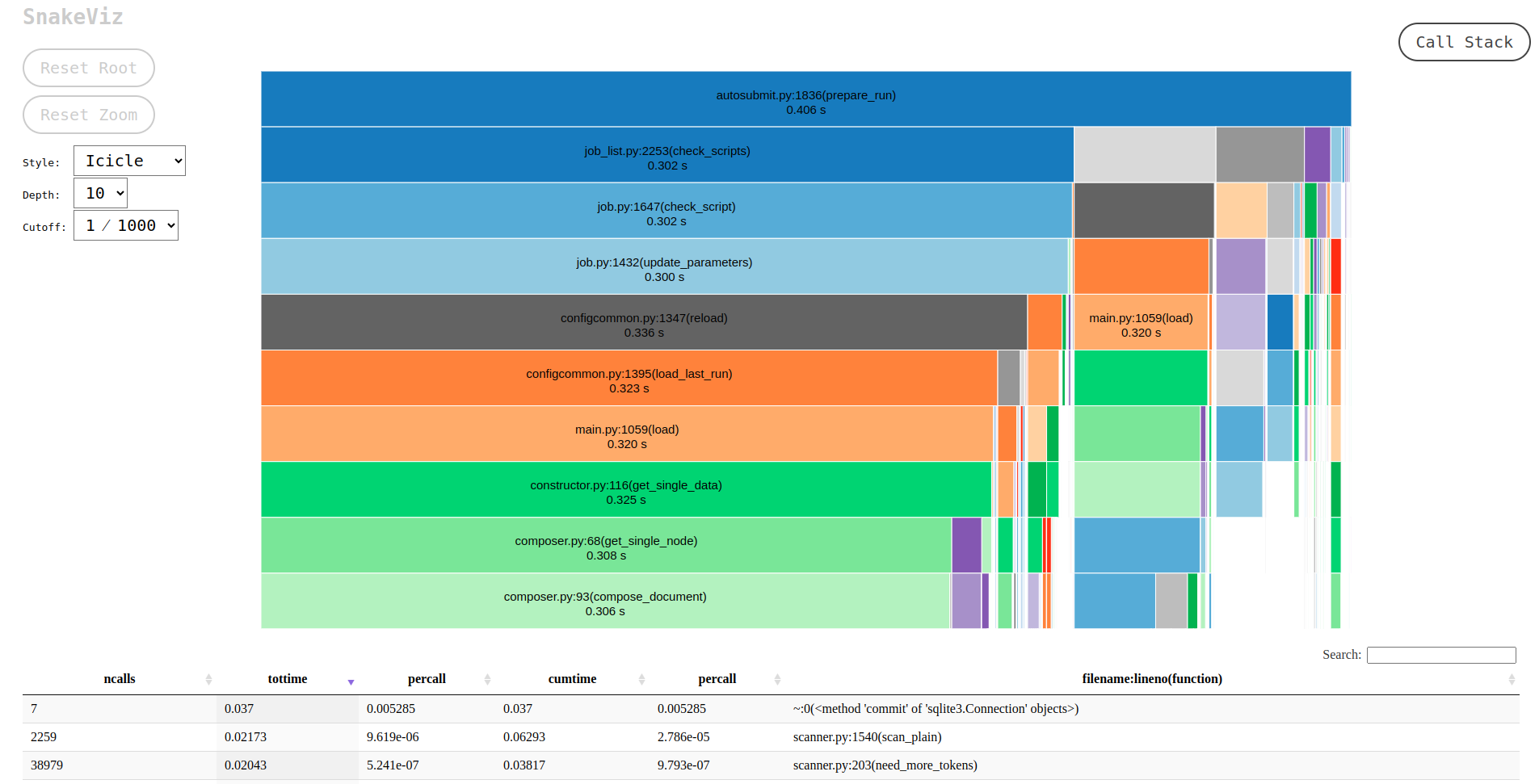
For more detailed documentation about the profiler, please visit this page.
How to get details about the experiment#
To get details about the experiment, use the command:
autosubmit describe {EXPID} {-u USERNAME}
EXPID is the experiment identifier, can be a list of expid separated by comma or spaces -u USERNAME is the username of the user who submitted the experiment.
It displays information about the experiment. Currently it describes owner,description_date,model,branch and hpc
Options:
usage: autosubmit describe [-h ] expid
expid experiment identifier
-u USERNAME, --user USERNAME username of the user who submitted the experiment
-h, --help show this help message and exit
Examples:
.. code-block:: bash
autosubmit describe cxxx autosubmit describe “cxxx cyyy” autosubmit describe cxxx,cyyy autosubmit describe -u dbeltran
How to monitor job statistics#
The following command could be adopted to generate the plots for visualizing the jobs statistics of the experiment at any instance:
autosubmit stats EXPID
EXPID is the experiment identifier.
Options:
usage: autosubmit stats [-h] [-ft] [-fp] [-o {pdf,png,ps,svg}] expid
expid experiment identifier
-h, --help show this help message and exit
-ft FILTER_TYPE, --filter_type FILTER_TYPE
Select the job type to filter the list of jobs
-fp FILTER_PERIOD, --filter_period FILTER_PERIOD
Select the period of time to filter the jobs
from current time to the past in number of hours back
-o {pdf,png,ps,svg}, --output {pdf,png,ps,svg}
type of output for generated plot
--hide, hide the plot
-nt --notransitive
prevents doing the transitive reduction when plotting the workflow
Example:
autosubmit stats cxxx
The location where user can find the generated plots with date and timestamp can be found below:
<experiments_directory>/cxxx/plot/cxxx_statistics_<date>_<time>.pdf
Console output description#
Example:
Period: 2021-04-25 06:43:00 ~ 2021-05-07 18:43:00
Submitted (#): 37
Run (#): 37
Failed (#): 3
Completed (#): 34
Queueing time (h): 1.61
Expected consumption real (h): 2.75
Expected consumption CPU time (h): 3.33
Consumption real (h): 0.05
Consumption CPU time (h): 0.06
Consumption (%): 1.75
Where:
Period: Requested time frame.
Submitted: Total number of attempts that reached the SUBMITTED status.
Run: Total number of attempts that reached the RUNNING status.
Failed: Total number of FAILED attempts of running a job.
Completed: Total number of attempts that reached the COMPLETED status.
Queueing time (h): Sum of the time spent queuing by attempts that reached the COMPLETED status, in hours.
Expected consumption real (h): Sum of wallclock values for all jobs, in hours.
Expected consumption CPU time (h): Sum of the products of wallclock value and number of requested processors for each job, in hours.
Consumption real (h): Sum of the time spent running by all attempts of jobs, in hours.
Consumption CPU time (h): Sum of the products of the time spent running and number of requested processors for each job, in hours.
Consumption (%): Percentage of Consumption CPU time relative to Expected consumption CPU time.
Diagram output description#
The main stats output is a bar diagram. On this diagram, each job presents these values:
Queued (h): Sum of time spent queuing for COMPLETED attempts, in hours.
Run (h): Sum of time spent running for COMPLETED attempts, in hours.
Failed jobs (#): Total number of FAILED attempts.
Fail Queued (h): Sum of time spent queuing for FAILED attempts, in hours.
Fail Run (h): Sum of time spent running for FAILED attempts, in hours.
Max wallclock (h): Maximum wallclock value for all jobs in the plot.
Notice that the left scale of the diagram measures the time in hours, and the right scale measures the number of attempts.
Custom statistics#
Although Autosubmit saves several statistics about your experiment, as the queueing time for each job, how many failures per job, etc.
The user also might be interested in adding his particular statistics to the Autosubmit stats report (`autosubmit stats EXPID`).
The allowed format for this feature is the same as the Autosubmit configuration files: INI style. For example:
COUPLING:
LOAD_BALANCE: 0.44
RECOMMENDED_PROCS_MODEL_A: 522
RECOMMENDED_PROCS_MODEL_B: 418
The location where user can put this stats is in the file:
<experiments_directory>/cxxx/tmp/cxxx_GENERAL_STATS
Hint
If it is not yet created, you can manually create the file: `expid_GENERAL_STATS` inside the `tmp` folder.
How to extract information about the experiment parameters#
This procedure allows you to extract the experiment variables that you want.
The command can be called with:
autosubmit report EXPID -t "absolute_file_path"
Alternatively it also can be called as follows:
autosubmit report expid -all
Or combined as follows:
autosubmit report expid -all -t "absolute_file_path"
Options:
usage: autosubmit report [-all] [-t] [-fp] [-p] expid
expid Experiment identifier
-t, --template <path_to_template> Allows to select a set of parameters to be extracted
-fp, --show_all_parameters All parameters will be extracted to a different file
-fp, --folder_path By default, all parameters will be put into experiment tmp folder
-p, --placeholders disable the replacement by - if the variable doesn't exist
Autosubmit parameters are encapsulated by %_%, once you know how the parameter is called you can create a template similar to the one as follows:
- **CHUNKS:** %NUMCHUNKS% - %CHUNKSIZE% %CHUNKSIZEUNIT%
- **VERSION:** %VERSION%
- **MODEL_RES:** %MODEL_RES%
- **PROCS:** %XIO_NUMPROC% / %NEM_NUMPROC% / %IFS_NUMPROC% / %LPJG_NUMPROC% / %TM5_NUMPROC_X% / %TM5_NUMPROC_Y%
- **PRODUCTION_EXP:** %PRODUCTION_EXP%
- **OUTCLASS:** %BSC_OUTCLASS% / %CMIP6_OUTCLASS%
This will be understood by Autosubmit and the result would be similar to:
- CHUNKS: 2 - 1 month
- VERSION: trunk
- MODEL_RES: LR
- PROCS: 96 / 336 / - / - / 1 / 45
- PRODUCTION_EXP: FALSE
- OUTCLASS: reduced / -
Although it depends on the experiment.
If the parameter doesn’t exists, it will be returned as ‘-’ while if the parameter is declared but empty it will remain empty
HPCQUEUE: bsc_es
HPCARCH: marenostrum4
LOCAL_TEMP_DIR: /home/dbeltran/experiments/ASlogs
NUMCHUNKS: 1
PROJECT_ORIGIN: https://earth.bsc.es/gitlab/es/auto-ecearth3.git
MARENOSTRUM4_HOST: mn1.bsc.es
NORD3_QUEUE: bsc_es
NORD3_ARCH: nord3
CHUNKSIZEUNIT: month
MARENOSTRUM4_LOGDIR: /gpfs/scratch/bsc32/bsc32070/a01w/LOG_a01w
PROJECT_COMMIT:
SCRATCH_DIR: /gpfs/scratch
HPCPROJ: bsc32
NORD3_BUDG: bsc32